Alternative Polyadenylation (APA) is Widespread in Eukaryotes:
Apart from a few cases, we do not understand why eukaryotic mRNAs require so many 3' end isoforms. Recent work based on the analysis of expressed sequence tags in tissues shows differential usage of polyadenylation sites (PAS), where ovary, retina, placenta and blood use proximal PAS sites, while bone marrow, uterus and brain prefer distal ones. However, it is not understood why APA is needed at a cell-type or tissue-specific level.
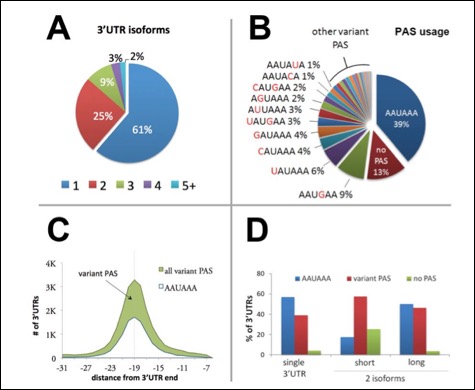
This figure shows A) 3'UTR isoform analysis: nearly half of worm protein-coding genes use multiple polyadenylation elements. B) The most abundant PAS element is AAUAAA, but other hexamers are also present C) The PAS element is invariably enriched at position -19 nt from the mRNA cleavage point. This chart shows the distribution of the ten most common PAS elements within the last 30 nucleotides of the mRNA of protein-coding genes. The vertical dashed lines indicate the -19 nucleotide point. D) Chart representing the preferred PAS element detected in 3'UTRs of genes with only one 3'UTR isoform and in the longest or shortest 3'UTR for genes with two 3'UTRs isoforms. While the canonical AAUAAA site is the preferred PAS signal element for the longer isoforms and for genes with only one isoform, the shorter isoforms are enriched with regions with no detectable PAS signal.
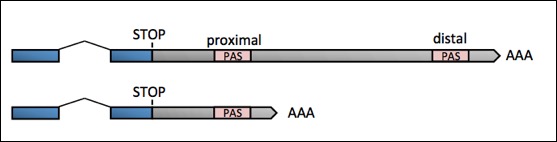
Cleavage and Polyadenylation dictates the Length of 3'UTRs and Generates APA:
CPSF and CstF are required for cleavage and polyadenylation of mature mRNAs:
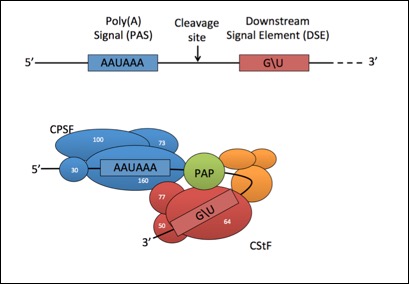
The choice between multiple PAS elements in the same 3'UTR is largely unknown:
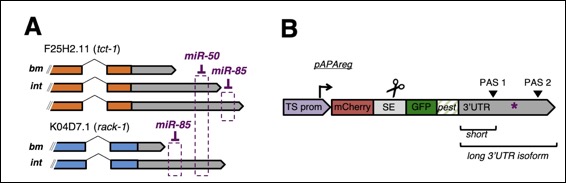
We are examining if the alternative 3'UTR isoforms are produced by changes in abundance of specific subunits of the cleavage complex, and if there are tissue-specific accessory factors that allow the production of specific 3'UTR isoforms. We address the first hypothesis by quantifying the relative abundance of the cleavage complex subunits in different tissues using qPCR. To address the second hypothesis, we are preparing sensor worm strains that sense the occurrence of 3'end cleavage of a variant or PAS-less 3'UTR and report it using mCherry visualization in a tissue-specific manner. The test 3'UTR isoform containing the variant element or PAS-less 3'UTR is fused to mCherry. When recombined in worms, the GFP will visualize the promoter activity (i.e. the presence of the mRNA), and the mCherry will report the occurred cleavage and polyadenylation of the tested 3'UTR. The silencing of any component of the cleavage machinery will result in the loss of mCherry expression, because the RNA is not polyadenylated and hence will be degraded.
We are preparing several different sensor strains corresponding to selected common variations of the canonical PAS element and representatives of the PAS-less 3'UTR. These strains express the 3'UTR in the corresponding tissue and are used in genome-wide RNA interference screens to identify genes involved in processing these variant and PAS-less elements.
Worms are particularly suitable for large-scale RNAi screens because the double-stranded RNA response can be easily induced by feeding available RNAi libraries. For each sensor strain we perform ~20,000 RNAi experiments, silencing every worm protein-coding gene and test for the loss of mCherry expression. Positive hits are re-screened, and genes that pass the second screen are studied in detail.