
We are specifically interested in the formation and biological role of the 3’ ends of genes known as 3’ Untranslated Regions (3’UTRs). These regions have recently
been subject to intense study, as they were found to be targets of various regulatory factors that recognize poorly characterized elements present in these regions, dosing gene expression.
One of the most intriguing results in the past years of research is that alternative polyadenylation (APA), a mechanism in which the same gene is processed with multiple ends of different lengths, is widespread in metazoans for reasons not yet understood.
We believe APA could represent a pervasive regulatory mechanism with potentially greater significance than what is currently accepted in the field. This mechanism involves the presence of shortened versions of a gene that lack the target sites for repressive elements or the presence of longer versions of the same gene that contain these target sites and are thus subject to regulation. This novel idea, which is the focus of our current research at ASU, could represent a paradigm shift in the transcription field. In my lab, I seek to fill this gap in understanding by characterizing this phenomenon.
My group is currently split into three areas of APA-focused research:
(1) Characterization of APA events in living organisms; how APA is achieved and regulated during metazoan development
In this research, we use the nematode C. elegans as a model system. Most APA research is currently conducted using cell lines, with most of these being cancer cells.
We believe that for such a fundamental process, an intact living organism such as C. elegans provides unique and highly precise insights into the biological implications of APA as a mechanism to modulate gene regulation, leading to a better understanding of such fundamental processes in humans. In addition, when compared to other metazoans, its gene model is currently the best annotated available, and the developmental lineage of each cell is extremely well characterized, allowing the link between APA and cell biology to be revealed more effectively than in other, more complex models.
Worms are also amenable for in vivo studies and can be easily manipulated in a high-throughput fashion with powerful genetic and molecular biology tools.
The main hypothesis of this research is that APA has a larger-than-suspected tissue-specific distribution, and genes may use it as a widespread regulatory mechanism that plays a role in establishing and maintaining cell identity at a tissue-specific level.
read more…
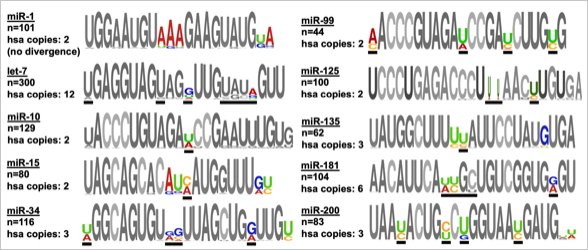
(2) Characterization of miRNA targets, their regulatory networks in cells, and their impact on human disease
MiRNAs are small, regulatory RNAs that pair with protein-coding genes, repressing their translation and limiting their activity. Because this recognition
elements are degenerated and small, miRNA targets are generally difficult to identify, thus the vast majority of miRNA targets are still unknown.
Defining these targets is the ultimate goal in the field of miRNA biology, as misregulation of miRNAs and their known target genes have been observed in almost all human diseases, including cancer, Alzheimer’s, and muscular disorders, suggesting a potential novel therapeutic approach for the development of future drugs. Here at ASU, my laboratory developed an innovative tool, 3’LIFE (Luminescent Identification of Functional Elements in 3’UTRs), which precisely defines these elements in genes. This high-throughput functional method is based on the well-known luciferase assay, and screens for the binding requirements of given miRNAs within a panel of 3’UTRs arrayed in 96-well plates. We recently used this assay to detect biologically important target genes, querying two breast cancer-relevant miRNAs, let-7c and miR-10b, to an array of ~400 test 3’UTRs.
read more… and read more…
(3) The role of APA in Duchenne muscular dystrophy (DMD)
DMD is an incurable, X-linked recessive genetic disorder characterized by progressive muscular degeneration. DMD is caused by the loss of function of dystrophin. This muscle cell membrane protein links the cytoskeleton with the muscle extracellular matrix and prevents cell membrane damage caused by repetitive muscle contractions. The absence of dystrophin leads to widespread consequences affecting multiple cell processes, ultimately leading to sarcolemma instability, apoptosis, and necrosis. We do not know how or when these cell autonomous processes are initiated and executed.
Recently, a new and very promising C. elegans DMD model became available, allowing for studying this disease in a simple, tractable organism. This strain lacks the worm homolog of the dystrophin gene (dys-1), and presents many features of the disease, such as a shortened lifespan and progressive muscle paralysis, which can be rescued by reintroducing the human dystrophin gene.
Because DMD is a progressive disease, our working hypothesis is that the loss of dystrophin activates unknown additive genetic changes at transcriptome and miRNA levels, which in turn use APA to control the severity of the disease.
read more…
