Graduate Students
Current

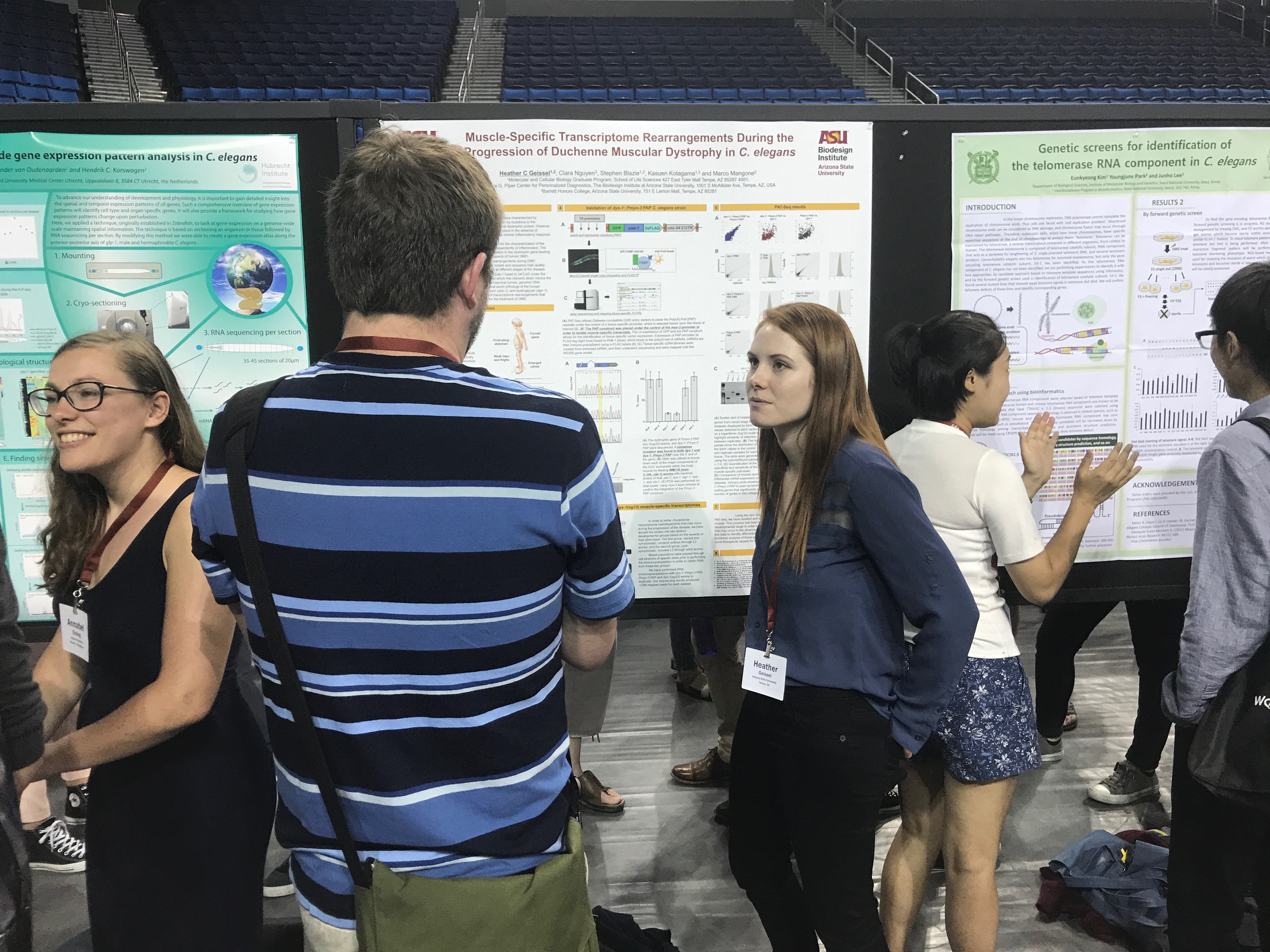
Heather Hrach (Geissel)
Graduate Student (2016-Current)Email: Heather.Geissel@asu.edu
Project: The goal of my project is to use C. elegans as a model organism to study Duchenne Muscular Dystrophy. This form of muscular dystrophy is caused by by mutations in the dystrophin gene, a muscular protein that links the internal cytoskeletal system of individual myofibers to structural proteins within the extracellular matrix. The specifics of my project will involve studying transcriptome and miRNAome rearrangements that occur in the absence of the dystrophin ortholog dys-1, in C. elegans . I will use two previously established worm strains that will allow for the tissue specific isolation of muscle from worms lacking dys-1, in effort to perform a functional analysis of differentially expressed genes in dys-1 dependent muscle degeneration.
Selected Publications:

Transcriptome changes during the initiation and progression of Duchenne Muscular Dystrophy in C. elegans
Human Molecular Genetics (accepted)

miRNA Profiling for Early Detection and Treatment of Duchenne Muscular Dystrophy.
Int. J. Mol. Sci. 2019, 20(18), 4638

Alternative polyadenylation directs tissue specific miRNA targeting in Caenorhabditis elegans somatic tissues
GENETICS (2017) https://doi.org/10.1534/genetics.116.196774

Anna Schorr
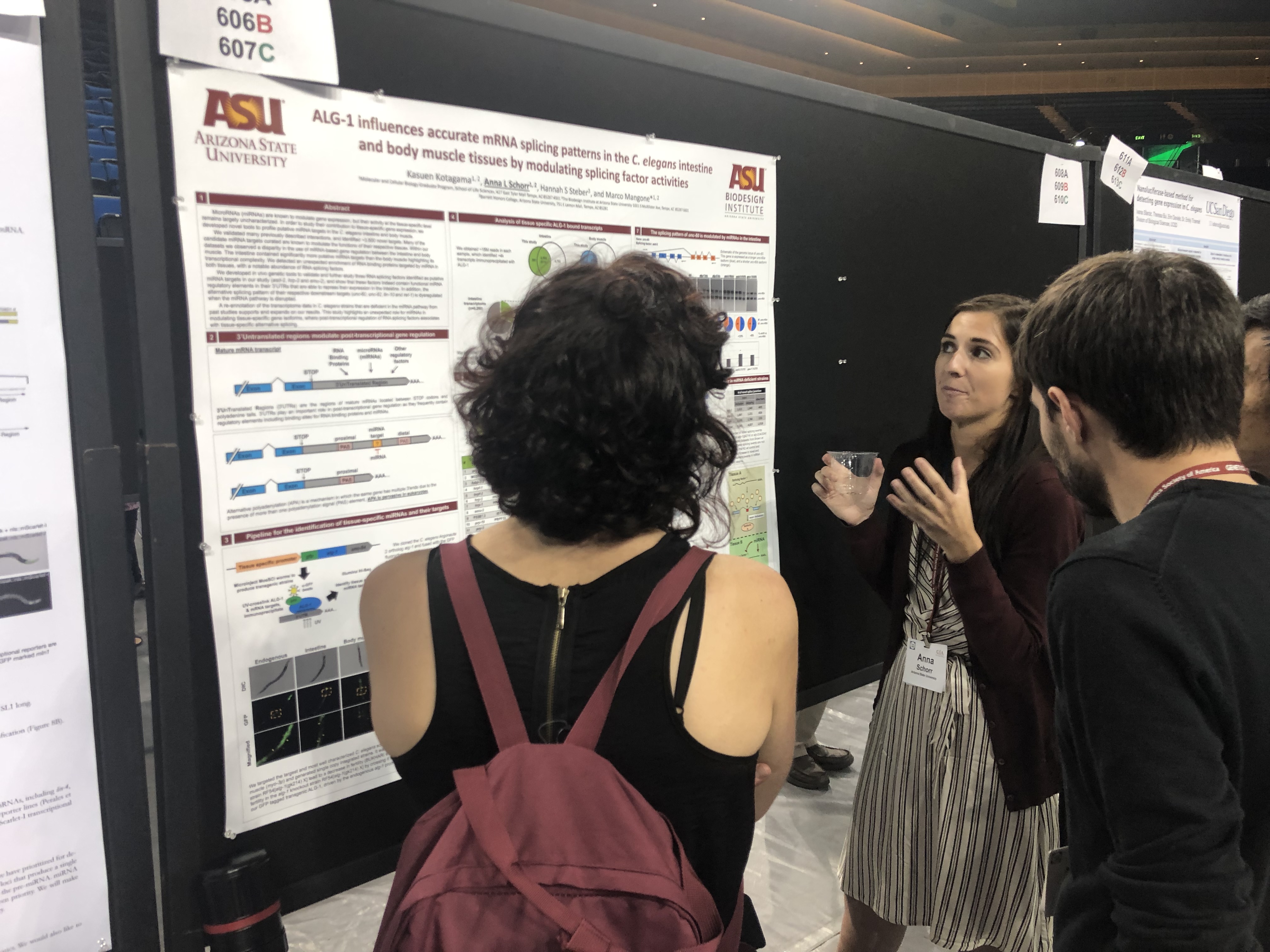
Graduate Student (2019-Current)Email: Anna.Schorr@asu.edu
Project: The objective of my research is to identify tissue-specific miRNA and miRNA targets and examine their role in gene regulation using the model organism C. elegans. Previous projects utilizing a GFP-tagged Argonaute protein immunoprecipitation approach demonstrated that there are distinct transcripts targeted by miRNA in the intestine and body muscle. I plan to expand this research by identifying mRNA targets in other tissues and cell types. However, more sensitive methods are required to isolate and identify specific miRNAs, especially in relatively small tissues comprised of few cells. To accomplish this, I am developing a technique to isolate nuclei in a tissue specific manner due to the relative ease of isolating nuclei instead of entire cells in an animal model. Additional applications of tissue- or cell-specific nuclei isolation will provide avenues for future research.
Selected Publications:

ALG-1 Influences Accurate mRNA Splicing Patterns in the Caenorhabditis elegans Intestine and Body Muscle Tissues by Modulating Splicing Factor Activities
GENETICS (2017) June 1, vol. 206 no. 2 757-774

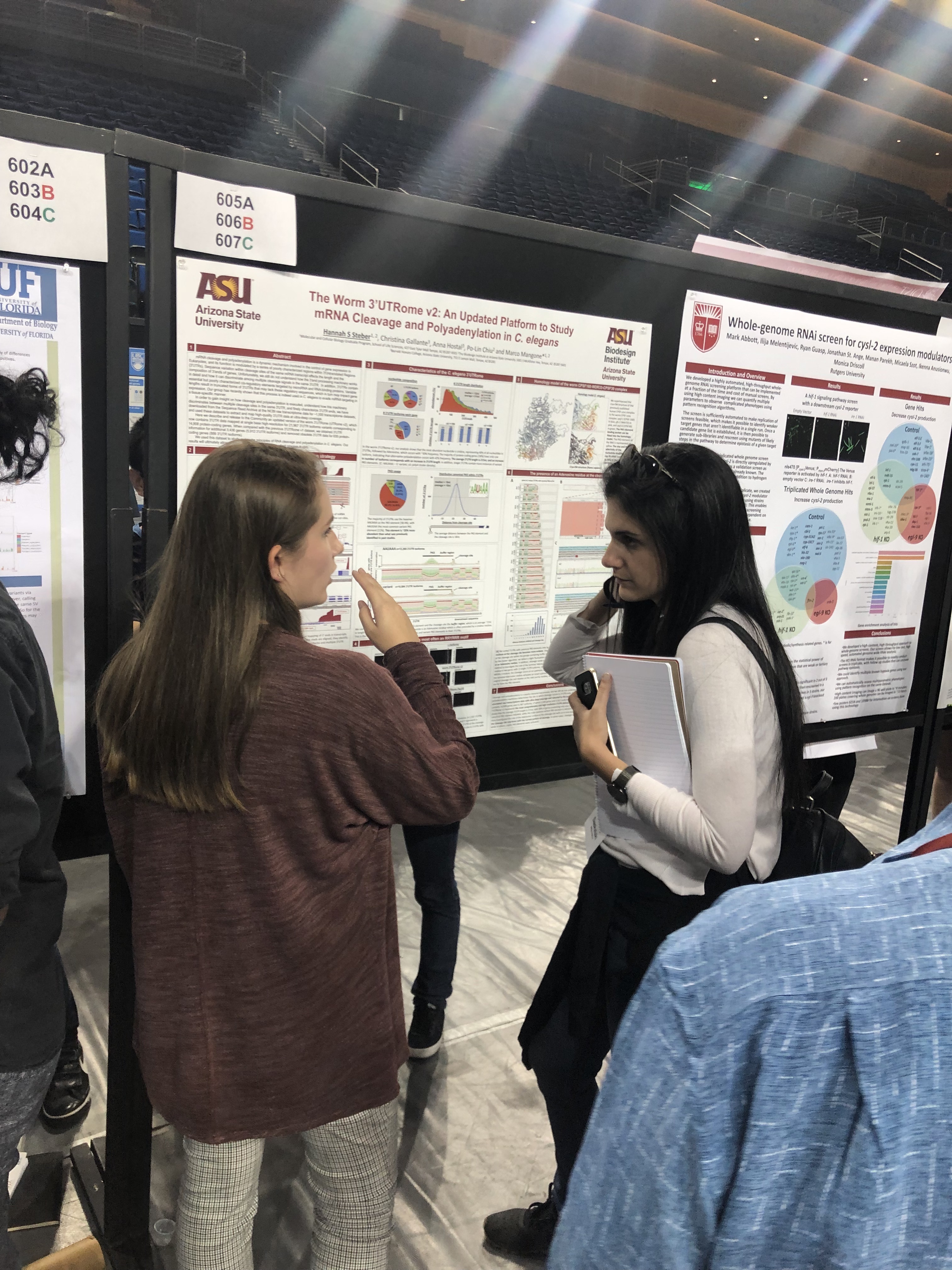
Hannah Steber
Graduate Student (2019-current)Email:Hannah.Steber@asu.edu

The C. elegans 3’-UTRome v2 resource for studying mRNA cleavage and polyadenylation, 3’-UTR biology, and miRNA targeting
Genome Research (2019) in press

ALG-1 Influences Accurate mRNA Splicing Patterns in the Caenorhabditis elegans Intestine and Body Muscle Tissues by Modulating Splicing Factor Activities
GENETICS (2017) June 1, vol. 206 no. 2 757-774
Past
![Biodesign Kasuen Kotagama 3[1]](files/biodesign-kasuen-kotagama-3005b1005d-2.png)
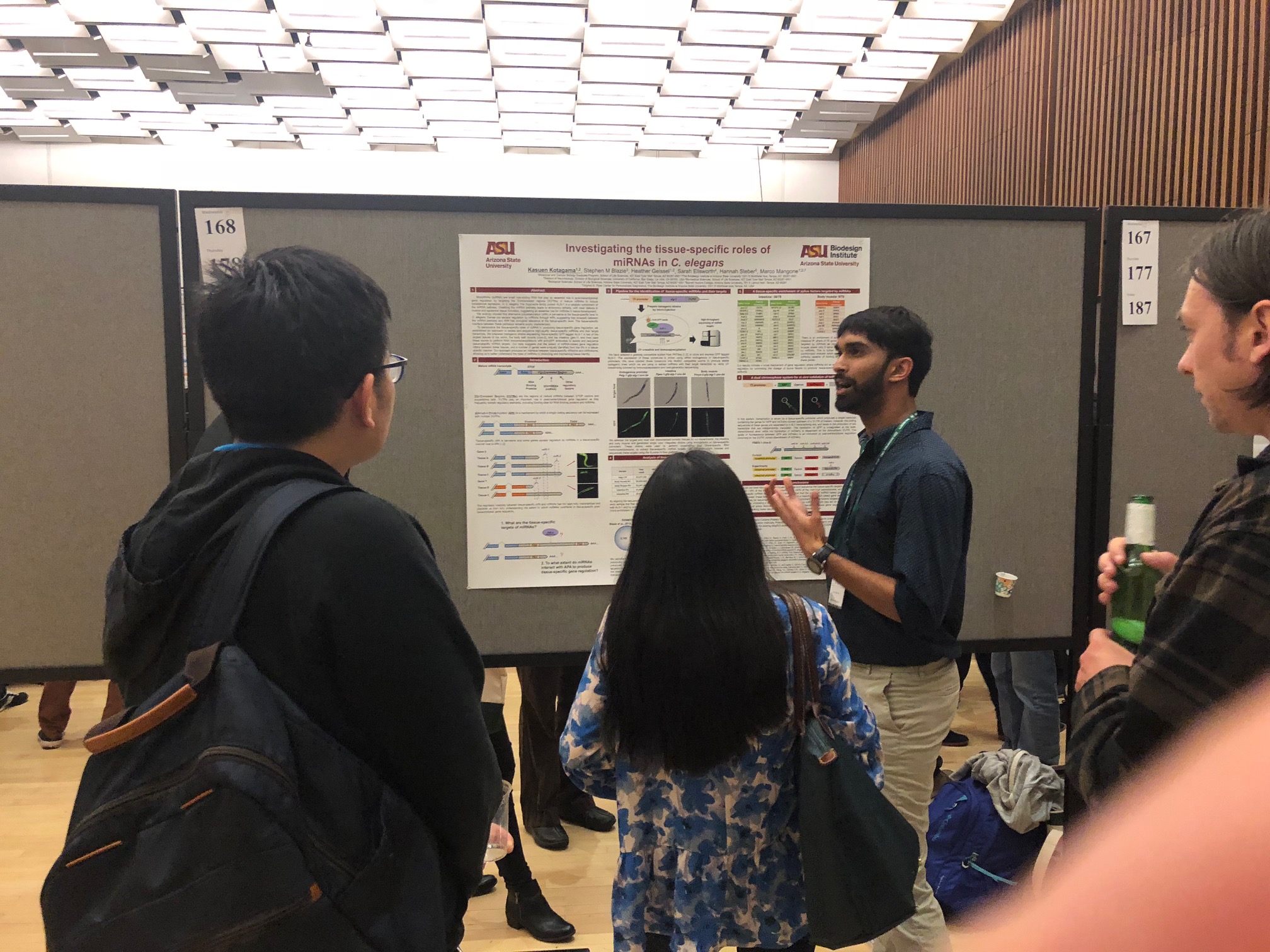
Kasuen Kotagama
Graduate Student (2014-Current) Email: Kasuen.Kotagama@asu.edu Project:The goal of my project is to integrate miRNA regulation with the mechanisms of 3′UTR cleavage and polyadenylation in C. elegans. Studies to date have explored 3′UTR cleavage and polyadenylation using in vitro biochemical techniques and cell lines. These studies have explored cleavage and polyadenylation using the canonical polyadenylation sequence (PAS) AAUAAA. However a majority of C. elegans 3`UTRs are processed using non-canonical PAS elements which suggests that in vivo cleavage and polyadenylation may be more complicated than was previously believed. I am currently developing a sensor strain of C. elegans, which can be used to study in vivo cleavage and polyadenylation at a tissue specific level. This sensor strain that we call the Cleavage and Polyadenylation Sensor Strain (CAPS) will be used to identify sequence elements and protein factors that are important for processing canonical and non-canonical polyadenylation sequences in C. elegans. Once developed, I also hope to use this system to perform a high-throughput RNAi screen targeted against RNA binding proteins in order to identify factors that play a role in mRNA cleavage and polyadenylation. I will also use this system to study tissue specific alternative polyadenylation and to identify factors that play a role in distinguishing amongst PAS elements.Kasuen Graduated in June 2019 and he is now a Post doctoral fellow in Dr. Katherine McJunkin Lab at the NIH (NIDDK).
Selected Publications:

ALG-1 Influences Accurate mRNA Splicing Patterns in the Caenorhabditis elegans Intestine and Body Muscle Tissues by Modulating Splicing Factor Activities
GENETICS (2017) June 1, vol. 206 no. 2 757-774

Evolutionary patterns of metazoan microRNAs reveal targeting principles in the human let-7 and miR-10 families
Genome Research (2017) 27: 53-63

A human 3′UTR clone collection to study post-transcriptional gene regulation
BMC Genomics (2015) 16:1036

miRNAs as Biomarkers in Chronic Myelogenous Leukemia
Drug Dev Res (2015) Aug 18. doi: 10.1002/ddr.21266

Detection of miRNA Targets in High-Throughput Using the 3'LIFE Assay
J Vis Exp (2015) May 25;(99):e52647. doi: 10.3791/52647

3′LIFE: a functional assay to detect miRNA targets in high-throughput
Nucleic Acids Res (2014) doi: 10.1093/nar/gku626

Stephen Blazie
Graduate Student (2012-2016)Email: Stephen.Blazie@asu.edu
Project: My goal is to integrate alternative polyadenylation to miRNA regulation in worms. I have taken a multi-pronged genomic approach directed toward refining the 3'UTRome by detecting tissue-specific 3'UTR isoform dynamics and their mechanisms of production. To identify the localization of tissue-specific 3' UTR isoforms, I am generating tissue specific mRNA libraries from C. elegans tissues coupling the mRNA tagging method with deep sequencing. I am preparing, and are currently sequencing, transcriptomes from MosSCI-derived stable transgenic worm lines driving expression of 3xFLAG-tagged Poly-A Binding Protein (pab-1) in eight tissues: muscle (myo-2, myo-3), nervous (unc-47, nmr-1), hypodermal (dpy-7, grd-10), and epithelial (ges-1, ajm-1). To further deconvolve the 3'UTRome, I am generating two transgenic worm lines expressing 3xFLAG-tagged worm orthologs for CPSF-160 (cpsf-1) and CstF-64 (cpf-2), that will be subjected to the HITS-CLIP approach to map their mRNA binding sites and study their contribution to APA. Results from these analyses represent the natural next step in improving the already unparalleled worm 3'UTRome annotation and will advance our insight of 3'UTR-mediated gene regulation.
Stephen Graduated in May 2016 and he is now a Post doctoral fellow in Dr. Yishi Jin Lab at UCSD.
Selected Publications:

Alternative polyadenylation directs tissue specific miRNA targeting in Caenorhabditis elegans somatic tissues
GENETICS (2017) https://doi.org/10.1534/genetics.116.196774

Comparative RNA-Seq analysis reveals pervasive tissue-specific alternative polyadenylation in Caenorhabditis elegans intestine and muscles
BMC Biology (2015) 13:4 doi:10.1186/s12915-015-0116-6)

Justin Wolter
Graduate Student (2012-2016)Email: Justin.Wolter@asu.edu
Project: My goal of my project in the Mangone Lab is to develop a high-throughput assay to identify miRNA targets throughout the genome. This assay, which we call Luminescent Identification of Functional Elements in 3'UTR's (3'LIFE), employs an innovative conjunction of biochemistry, genetics, and high-throughput genomics to characterize gene networks targeted by miRNAs. Current approaches designed to identify miRNA targets are limited by issues of scalability and resolution. To address the issue of resolution, the 3'LIFE is based on the well-characterized dual-luciferase and PAR-CLIP assays, identifying genes regulated by a given miRNA and validating targeted elements at single base-pair resolution in tandem. To overcome the limitation of scalability, we have adapted 3'LIFE to be compatible with the high-throughput robotics here at the Biodesign Institute, performing cloning, cell transfections, and luciferase assays in 96 well plates. Using breast cancer as a model system, we are focusing on two well characterized cancer-relevant miRNAs: one a tumor suppressor that is frequently deleted in various cancers, and one that is frequently overexpressed in metastatic and highly invasive breast tumors. To optimize the 3'LIFE assay we are screening a pilot dataset of 300 human 3'UTRs belonging to cancer-relevant genes, and testing these for targeting by the given miRNAs.
Justin Graduated in Oct 2016 and he is now a Post doctoral fellow in Dr. Mark Zylka Lab at UNC.
Selected Publications:

Evolutionary patterns of metazoan microRNAs reveal targeting principles in the human let-7 and miR-10 families
Genome Research (2017) 27: 53-63

Differential expression of conserved and novel microRNAs during tail regeneration in the lizard Anolis carolinensis
BMC Genomics (2016) 17:339

A human 3′UTR clone collection to study post-transcriptional gene regulation
BMC Genomics (2015) 16:1036

Detection of miRNA Targets in High-Throughput Using the 3'LIFE Assay
J Vis Exp (2015) May 25;(99):e52647. doi: 10.3791/52647

3′LIFE: a functional assay to detect miRNA targets in high-throughput
Nucleic Acids Res (2014) doi: 10.1093/nar/gku626