Luminescent Identification of Functional Elements in 3'UTRs: 3'LIFE
The 3'LIFE is an adaptation of the well-characterized dual luciferase reporter assay, and performed in high-throughput (HT). 3'LIFE rapidly identifies functional targets of a given miRNA in a panel of thousands of test 3'UTRs in co-transfection experiments.
3'LIFE uses two specially designed vectors to express the test 3'UTRs (pLIFE-3'UTR) and the miRNA (pLIFE-miRNA)
ORDER THE 3'-LIFE VECTORS HERE and HERE.
DETAILED 3'-LIFE PROTOCOL HERE
EXCEL SPREADSHEETS FOR 3'LIFE ASSAY HERE and HERE
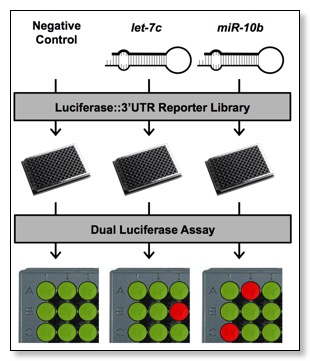
Currently the feasibility of high-throughput dual luciferase assays is limited by high costs associated with transfection and luciferase assay reagents, and the lack of a publicly available human 3'UTR library. Furthermore, genome-wide screens for miRNA targets are challenged by the need for appropriate high-throughput technologies and pipelines. To overcome these limitations, we have designed each step of the 3'UTR cloning pipeline and the 3'LIFE assay to be highly automated. PCR, cloning, plasmid DNA preparation, cell culture, transfection, and luciferase assays are performed in 96-well format using multi-channel micropipettes, dedicated liquid handling robots, and other high-throughput instrumentation. We have spent significant effort on cost reduction. We have optimized construct transfections in HEK293T cells, which are efficiently transfected in reusable 96-well nucleofection plates, and developed a series of buffers for use with the nucleofection hardware, virtually eliminating the cost of consumable transfection reagents. We also reduced the cost of the luciferase assay ~95% by creating in-house luciferase buffers. These cost-effective strategies will permit 3'UTR library expansion, additional screens for miRNA targets, and significantly increase the scope of data obtained by this project.
As a result of these developments, 3'LIFE overcomes the current limitations of scalability and resolution, allowing for the first time the systematic and unbiased identification of miRNA targets. Importantly, 3'LIFE conveys several advantages over current methods. First, 3'LIFE is a comprehensive screen because it identifies all functional elements present in each 3'UTR that are targeted by the query miRNA. Second, 3'LIFE is unbiased, since it probes one interaction at a time and does not rely on prior assumptions about target genes. Third, the luciferase reporter requires direct inhibition of the 3'UTR in the presence of the miRNA, eliminating indirect regulation frequently detected by mRNA and protein based approaches. Fourth, 3'LIFE discriminates between functional and non-functional miRNA target sites (see preliminary results below). Lastly, 3'LIFE can be adapted to detect other functional elements in 3'UTRs that are targeted by non-coding RNAs and RNA binding proteins. 3'LIFE provides rapid detection and initial validation of direct miRNA/3'UTR target interactions at a scale not possible with current methods.
