POLYA TRANSCRIPTS SEQUENCING: PAT-SEQ
mRNA-tagging uses tissue-specific promoters to drive expression of a FLAG epitope-tagged poly-A binding protein (PABP) which specifically binds to the poly(A)-tail of mRNAs, followed by crosslinking and immunoprecipitation of tissue-specific mRNAs.
Prior to our work mRNA-tagging was coupled with low-resolution platforms, such as microarrays and tiling arrays, that lack the sensitivity required to detect lowly expressed transcripts, pinpoint exome changes, and map 3’UTRs at single base resolution. To improve upon its sensitivity and specificity, we made several key changes to the original protocol:
1) We added three tandem FLAG-epitope (3xFLAG) tags instead of one, to improve the efficiency of the FLAG pull-down.
2) In the original mRNA-tagging protocol the FLAG-tagged PABP construct is selectively expressed as extra chromosomal arrays, which are unstable, often leading to mosaic expression patterns. We instead opted for the widely used Mos-1 single copy insertion (MosSCI) technology 36, which stably incorporates the construct of interest in the worm genome.
3) We adopted a novel strategy to prepare the tissue-specific cDNA libraries that relies on linear amplification of mRNAs, which minimizes the quantification error rate due to limited starting material and provides high-quality transcriptome and 3'end data in the same experiment 37.
4) We replaced the microarray step with Next-Gen sequencing (HiSeq) to improve data resolution. We call this updated method polyA-tagging and sequencing (PAT-Seq).
Download the protocol
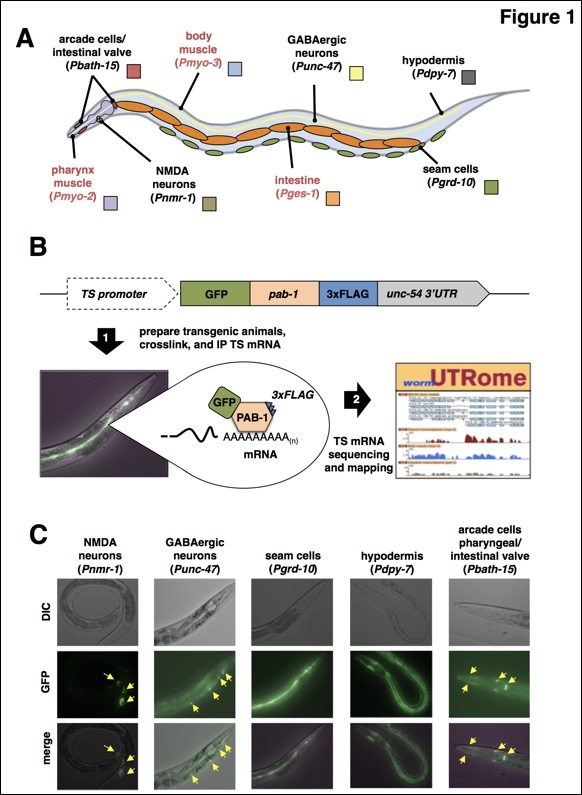
Figure 1: So far we have used PAT-Seq to sequence tissue specific transcriptomes from arcade and intestinal valve cells, NMDA-type neurons, GABAergic neurons, seam cells, and hypodermis. A) Diagram with the anatomical location of tissues we have profiled in this study. Red labels mark tissues that our group has previously profiled (Blazie et al., 2015), and that have been re-mapped in this study and used in the analysis. B) Overview of the PAT-Seq approach. The tissue specific expression of the Poly(A)-Pull cassette containing GFP fused to 3xFLAG-tagged pab-1 is achieved through usage of selected tissue-specific promoters. Transgenic worm lines that express this construct are then prepared and grown in liquid culture, followed by crosslinking, lysis and immunoprecipitation of 3xFLAG-tagged PAB-1 complexes. The tissue specific mRNA extracted from the IP is then used to prepare cDNA libraries for next generation sequencing and transcriptome mapping, which is stored in the UTRome.org database (Mangone et al., 2008). C) Examples of fluorescent images of worms expressing the Poly(A)-Pull cassette in the tissue profiled in this study. Yellow arrows mark small cells expressing the construct.
