miRNA Target Validation
3’LIFE Assay
The system, that we have named 3'-LIFE (Luminescence-based Identification of Functional Elements in 3'UTRs), overcomes the limitations of scalability and resolution of current methods, allowing the systematic and inexpensive mapping of regulatory elements in 3'UTRs.
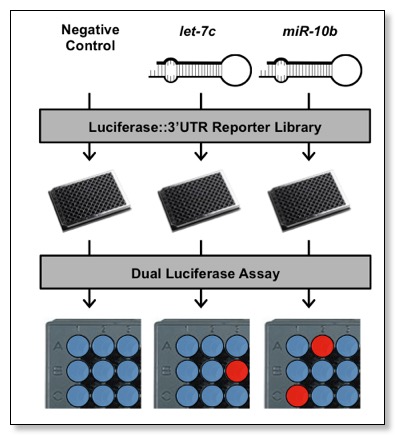
3'-LIFE is an adaptation of the well characterized dual luciferase reporter assay, but is scaled up and performed in high-throughput using 96-well plates handled by dedicated robots, together with robust bioinformatic analysis and wet-bench approaches. A major advantage of 3'LIFE is that it is a functional assay, meaning that the elements are detected and validated at the same time. The 3'LIFE assay is based on the dual-luciferase assay, a sensitive, rapid, and easily scalable assay to detect fluctuations in protein expression.
The dual luciferase assay relies on the fusion of a test 3'UTR to a luminescent reporter gene. Targeting and translational repression of the test 3'UTR by a probe miRNA is identified by a decrease in the luciferase::3'UTR signal (Firefly luciferase) relative to a second normalization signal (Renilla luciferase). 3'LIFE is designed to rapidly identify functional targets of a given miRNA in a panel of hundreds of test 3'UTRs in co-transfection experiments. The luminescence of each luciferase protein is measured sequentially in each well, and used to quantify translation of the reporter::3'UTR. Putative miRNA targets (red spots) are assigned when protein levels change in response to cotransfection with a miRNA compared to a negative control (blue spots).